Case Study: Protein Engineering via Mutagenesis and Affinity Maturation
August 5, 2021
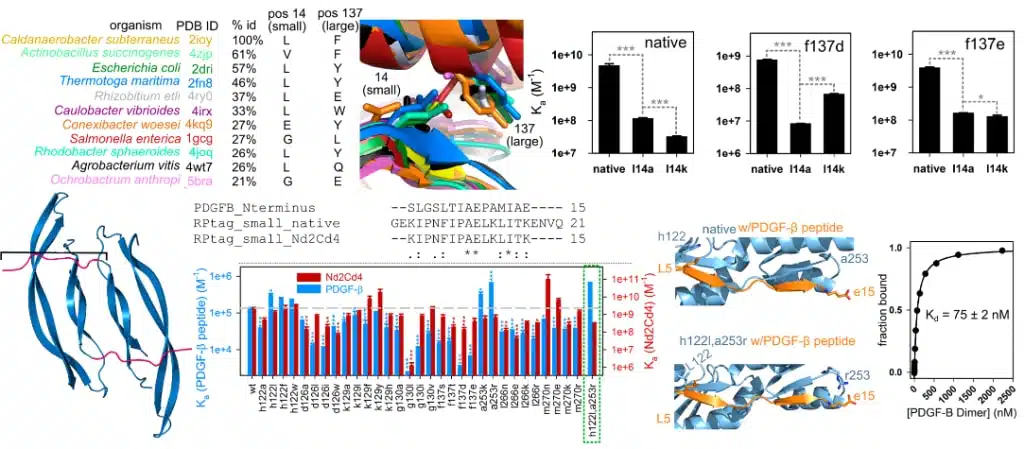
In 2018, we published an article highlighting the creation of a generalizable protein binding scaffold and epitope tag that is orally bioavailable called RPtag. RPtag is based on a ribose-binding protein isolated from an extremophile, awarding it expansive stability across highly variable environmental conditions. It was rationally engineered by splitting an irregular C-terminal pair of beta sheets, generating large and small fragments with high affinity and specificity to each other. We conducted directed mutagenesis to manipulate RPtag specificity by generating 31 mutants of RPtag (large) with a focus on positions proximal to nonconserved residues between RPtag (small) and PDGF-β, a clinically relevant growth factor. Dissocation constant measurements revealed two mutations that improved PDGF-β affinity but not the RPtag (small) sequence; the double mutant variant demonstrated superior affinity, enriching PDGF-β specificity 74x.
This is an example of our ability to optimize binding for novel proteins using structure guided rational mutagenesis. Our comprehensive understanding of the molecular mechanisms driving biological function is critical for the discovery of contemporary compounds, and a substantial advantage of Ichor workflows.
Biochemistry 2018, 57, 21, 3036–3049
Publication Date: May 3, 2018 https://doi.org/10.1021/acs.biochem.8b00170