Structural Biology Techniques for Protein Analysis and Characterization
Ichor’s structural biology services provide insights into the three-dimensional structures of biomolecules and their dynamics, helping you understand their biological function and developing targeted therapies.
- Protein Expression: Techniques that enable the production of sufficient proteins for structural analysis, through controlling the expression of proteins, you can study their structure, function, and interactions.
- Protein Purification: Essential for understanding protein function, drug discovery, and the development of new therapeutic agents, protein purification involves isolating a specific protein from a complex mixture of other biomolecules, in order to obtain a pure sample of the protein for further analysis.
- Protein Crystallisation: Crystallization of proteins arranges the molecules in a repetitive lattice. This allows for the determination of the three-dimensional structure of the protein at atomic resolution by using techniques such as X-Ray Crystallography.
- Biophysical Assays: Surface Plasmon Resonance (SPR), Isothermal Titration Calorimetry (ITC), and Thermal Shift Assay (TSA) examine the physical and chemical properties of proteins and their interactions. Combined with the support of structural biology services, these assays aid in characterizing protein structure, stability, dynamics, and binding interactions.
- X-Ray Crystallography: X-Ray Crystallography: Determine the three-dimensional structure of proteins. The target protein is screened for formation of crystals under varying physicochemical parameters (Example: pH, Temperature, concentration of salts etc.). The structures of the proteins are determined from the electron density maps computed from the X-ray diffraction patterns of the crystals.
- Nuclear Magnetic Resonance (NMR): NMR spectroscopy studies structure, dynamics, and molecular interactions of proteins in solution, in order to investigate protein-ligand interactions, protein folding, and conformational changes.
Structure Determination with Structural Biology Services
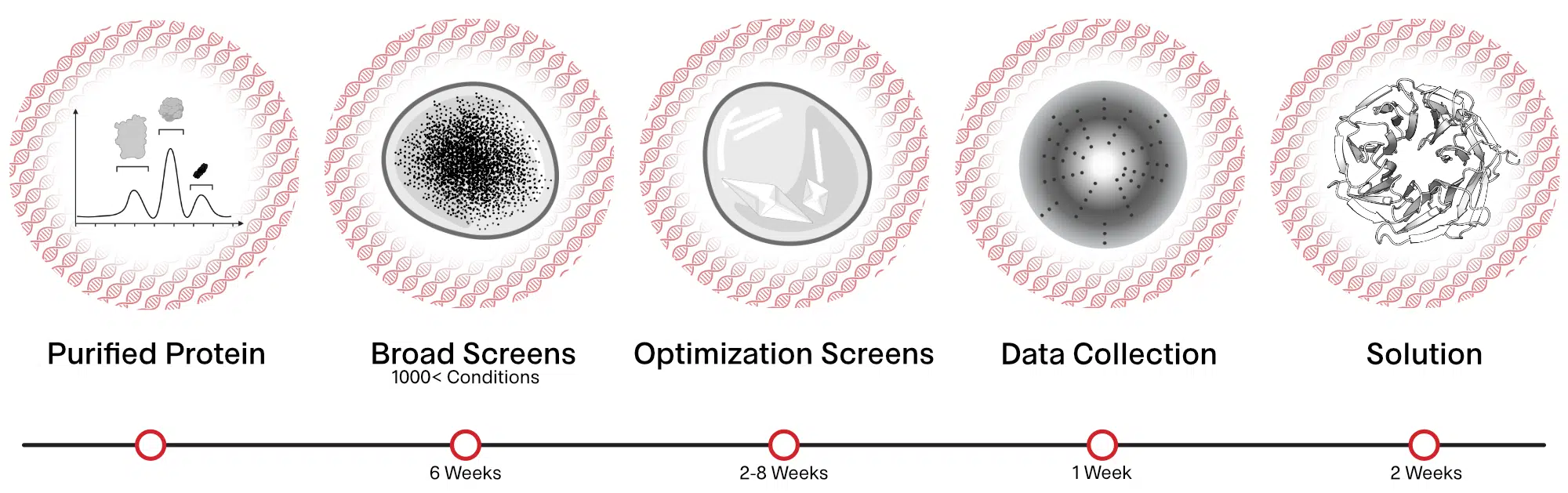
Purified and monodisperse protein preparations are screened for conditions that provide large enough crystals (>50 microns). These crystals are frozen in liquid nitrogen and analyzed at synchrotron sources to screen for crystals showing high resolution diffraction. The diffraction data is processed and a model of the protein at atomic resolution is obtained.
Primary Crystal Screening
Identification of promising crystallization conditions for protein of interest using 800-1000 conditions.
Optimization Screens
Optimize promising conditions identified from primary screens to obtain crystals.
Protein Modification
If promising conditions are not identified with the intact preparations of proteins, then the protein can be modified by either in situ proteolysis or lysine methylation. The modified protein can then be screened.
Protein-Ligand Complexes
Protein-ligand complexes, key focus of our structural biology services, can be collected through two main methods, by crystallising the protein in the presence of the ligand, or through soaking the ligand with apo-protein crystals.
X-ray Data Collection
Crystals obtained from the screens are frozen in liquid nitrogen and exposed to x-rays at synchrotron sources and the diffraction data is collected.
Structure Determination
X-ray diffraction data obtained are processed to obtain an electron density map. The electron density map together with the known protein sequence is used to obtain the three-dimensional structure of the protein.
Tailored Assays for Targeted Discovery
Our structural biology services offer comprehensive solutions for researchers and drug discovery teams to study and discover the biomolecular structures of protein and use this information to breakthrough scientific advancements.
Assay Development and Screening: Customized assay development and screening services tailored to your needs, facilitating target identification, validations, and compound screening. With our capacity for high-throughput techniques to ensure efficient screening of compound libraries, the discovery of novel leads and therapeutic candidates is accelerated with our structural biology services.
Fragment-Based Lead Discovery: Our structural biology services specialize in fragment-based lead discovery, a method that identifies small molecules with therapeutic potential, by screening a diverse collection of low molecular weight compounds, fragment-based lead discovery can determine the specific fragments that bind to the target protein of interest. Aiding in optimization, and leading to the development of potential drug candidates, our structural biology services will contribute to the process of hit identification and validation.
Instrumentation
We utilize state-of-the-art instrumentation to support our structural biology services, including:

Formulatrix NT8 Crystallization Robot
The NT8 robot can be used to set up sitting drop crystallization plates under 96 different conditions. This can be used to screen 3 different protein samples on a single crystallization plate.

Formulatrix Rock Imager 1000
The Rock Imager 1000 provides automated imaging of crystallization plates on a pre-programmed schedule.
Frequently Asked Questions
Answers to some common questions on protein structure determination, as part of our structural biology services.
The primary requirement for crystallizing a protein or protein complex is pure protein. If diffracting crystals are obtained, a known structure of a homologous protein would facilitate structure determination.
Primary crystallization screening involves incubating the protein under conditions varying different physicochemical parameters such as concentration of precipitants, concentration of salts, pH, and temperature. We start by screening the protein against 800-1000 popular crystallizing conditions.
Optimization screening involves varying physicochemical parameters around the promising conditions that have been identified from the primary screens. For example, if 20% PEG 8000 and 01. M Tris-HCl (pH 7.5) are identified as promising conditions that produce crystals, then optimization screens would be set up varying the PEG 8000 from 10 – 30% and the pH would be varied from 7.0 – 8.0.
Primary crystallization screening would require about 2-3 mg of purified protein. The protein should be 95-98 % pure as analyzed on Coomassie-blue stained gel. Another 2-3 mg of protein would be required for optimization screens and for growing crystals that will be sent to the synchrotron beamlines for data collection.